
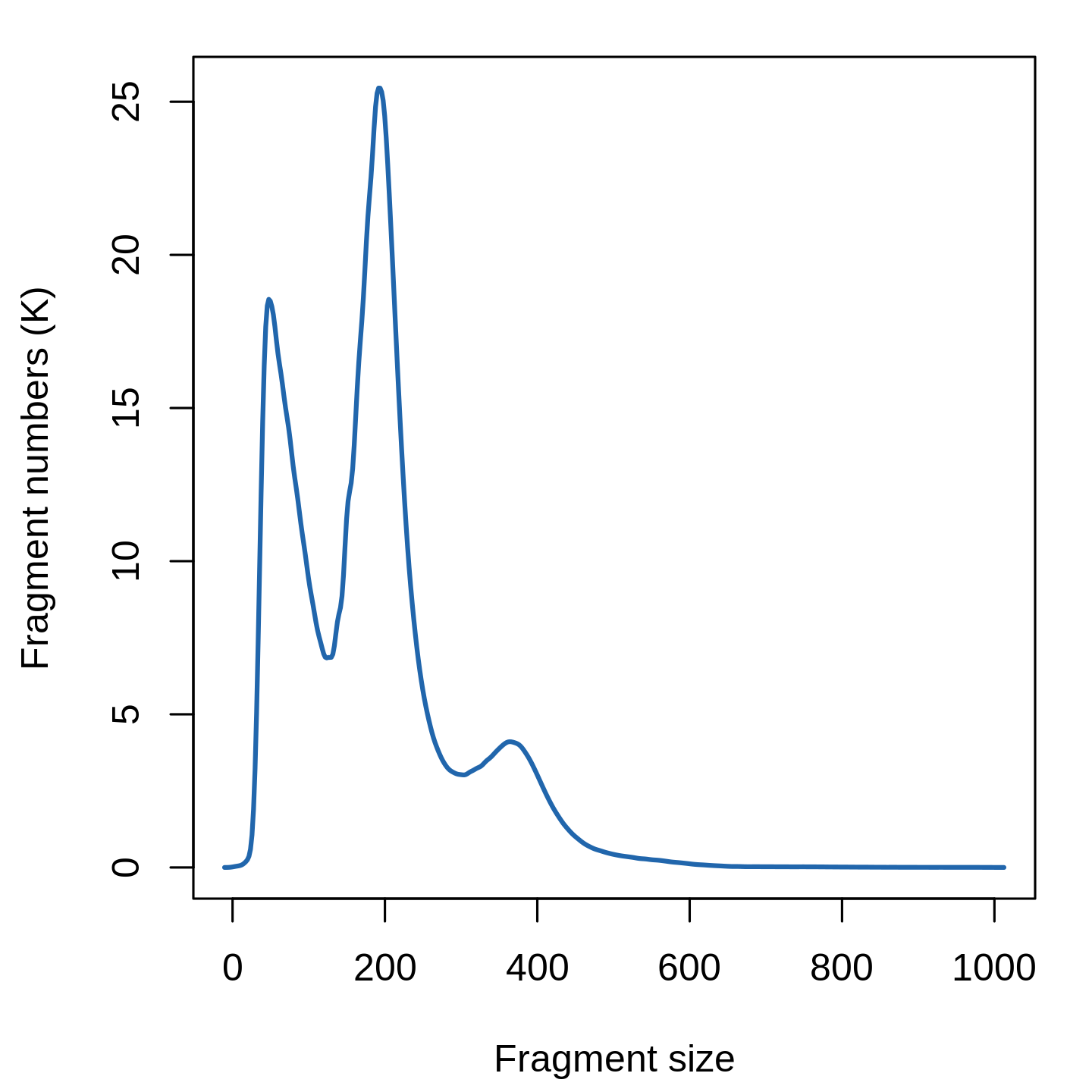
Here, we will all features, though we note that we see very similar results when using only a subset of features (try setting min.cutoff to ‘q75’ to use the top 25% all peaks), with faster runtimes. Instead, we can choose to use only the top n% of features (peaks) for dimensional reduction, or remove features present in less than n cells with the FindTopFeatures() function. This is a two-step normalization procedure, that both normalizes across cells to correct for differences in cellular sequencing depth, and across peaks to give higher values to more rare peaks.įeature selection: The low dynamic range of scATAC-seq data makes it challenging to perform variable feature selection, as we do for scRNA-seq. Normalization: Signac performs term frequency-inverse document frequency (TF-IDF) normalization. Normalization and linear dimensional reduction Note that the last three metrics can be obtained from the output of CellRanger (which is stored in the object metadata), but can also be calculated for non-10x datasets using Signac (more information at the end of this document). elegans (ce10) are included in the Signac package. ENCODE blacklist regions for human (hg19 and GRCh38), mouse (mm10), Drosophila (dm3), and C. Cells with a high proportion of reads mapping to these areas (compared to reads mapping to peaks) often represent technical artifacts and should be removed. Ratio reads in genomic blacklist regions The ENCODE project has provided a list of blacklist regions, representing reads which are often associated with artefactual signal. Note that this value can be sensitive to the set of peaks used. Cells with low values (i.e. <15-20%) often represent low-quality cells or technical artifacts that should be removed. Cells with extremely high levels may represent doublets, nuclei clumps, or other artefacts.įraction of fragments in peaks: Represents the fraction of all fragments that fall within ATAC-seq peaks. Cells with very few reads may need to be excluded due to low sequencing depth. Total number of fragments in peaks: A measure of cellular sequencing depth / complexity. We can compute this metric for each cell with the TSSEnrichment() function, and the results are stored in metadata under the column name TSS.enrichment. Poor ATAC-seq experiments typically will have a low TSS enrichment score. The ENCODE project has defined an ATAC-seq targeting score based on the ratio of fragments centered at the TSS to fragments in TSS-flanking regions (see ). Transcriptional start site (TSS) enrichment score. We calculate this per single cell, and quantify the approximate ratio of mononucleosomal to nucleosome-free fragments (stored as nucleosome_signal) Nucleosome banding pattern: The histogram of DNA fragment sizes (determined from the paired-end sequencing reads) should exhibit a strong nucleosome banding pattern corresponding to the length of DNA wrapped around a single nucleosome. As with scRNA-seq, the expected range of values for these parameters will vary depending on your biological system, cell viability, and other factors. We currently suggest the following metrics below to assess data quality.

We can now compute some QC metrics for the scATAC-seq experiment.

We start by creating a Seurat object using the peak/cell matrix and cell metadata generated by cellranger-atac, and store the path to the fragment file on disk in the Seurat object: More information about the fragment file can be found on the 10x Genomics website or on the sinto website. However, the advantage of retaining this file is that it contains all fragments associated with each single cell, as opposed to only fragments that map to peaks. It is a substantially larger file, is slower to work with, and is stored on-disk (instead of in memory).
#Posorted bam file format 10x scatac full#
This represents a full list of all unique fragments across all single cells.
You can find more detail on the 10X Website.įragment file. Each value in the matrix represents the number of Tn5 integration sites for each single barcode (i.e. a cell) that map within each peak. However, instead of genes, each row of the matrix represents a region of the genome (a peak), that is predicted to represent a region of open chromatin. This is analogous to the gene expression count matrix used to analyze single-cell RNA-seq. When pre-processing chromatin data, Signac uses information from two related input files, both of which can be created using CellRanger:


 0 kommentar(er)
0 kommentar(er)
